Who killed the unprotected RNA molecule?
Oeiras, 07.05.2012
Small non-coding RNA molecules that do not encode any protein are no longer regarded as junk. In fact, they play an active role determining how genes are expressed inside de cell. Now researchers from the Control of Gene Expression Lab have uncovered an alternative regulatory pathway in the dynamic RNA metabolism in bacteria, identifying the enzyme responsible for the degradation of unprotected small RNAs. The work is published in the April issue of the RNA journal.
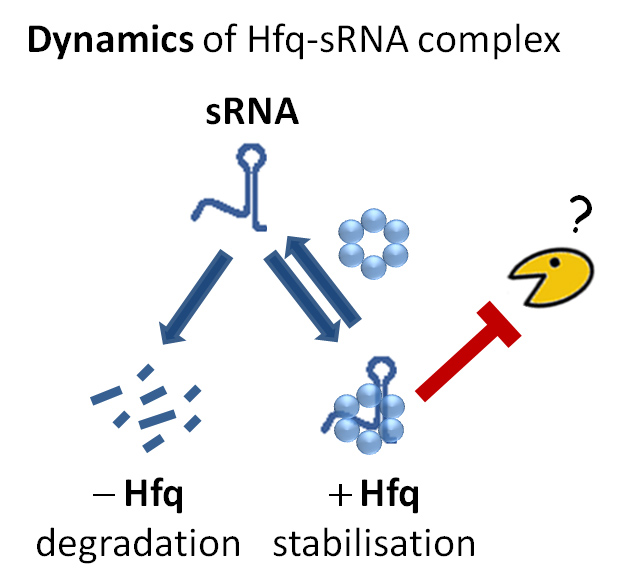
Bacterial small RNAs are dynamically associated to a protein called Hfq. When not protected by Hfq, small RNAs are rapidly degraded but the enzyme responsible for this degradation process was unknown. Considering all RNA degrading enzymes (ribonucleases) as suspects, researchers removed them one by one and looked at the effect on small RNA degradation. Just like murder suspects can be ruled out through their alibis, if the enzyme was not involved in the crime, removing it would not make any difference. On the other hand, inactivating the guilty enzyme would stop degradation. With this strategy, Polynucleotide Phosphorylase (PNPase) was identified as the culprit enzyme.
Before this work, the RNA community believed that a different type of enzyme (RNase E) was involved in unprotected small RNA degradation. The major difference between PNPase and RNase E the way they chop the RNA molecule, starting from its extremities (PNPase) or attacking its middle (RNase E). Researchers now believe that small RNA degradation by PNPase and the counter protection offered by the Hfq protein are “far more common in the living world than was previously envisaged”.
Original Article
The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq
José M. Andrade, Vânia Pobre, Ana M. Matos1 and Cecília M. Arraiano







