Structural Biology at IBN
Almost half of all known enzymes (40 to 47%) associate with a particular metal to function. In all of these, the metal center(s) are essential for catalysis, electron transfer and/or play a crucial role in ensuring stability and structural properties. Metalloproteins are well represented in metal-respiring organisms: Multiheme cytochromes (MHC) are the core proteins that most often guarantee the electron flow between solid electron donors/or acceptors and the inner-cell metabolism; Iron-sulfur proteins (such as HiPIPs) are usually found as the electron carriers between MHCs. Flavoproteins and 2Fe-2S proteins participate in pathways guaranteeing the high cellular iron uptake requirements and then, proteins (such as CcmI, and other maturation system proteins) are vital for ensuring the proper maturation and folding of metal-containing enzymes.
The three dimensional structure of metalloproteins is a key piece of information for understanding their function and role in physiological pathways, and is also essential for designing variants with desired properties. These designed metalloproteins can have improved stability, faster reaction kinetics, reduction potentials suitable for different reactions, or improved interaction with alternative physiological and non-physiological partners.
At the Inorganic Biochemistry Laboratory, we determine the structure of these metalloproteins either by X-Ray Crystallography (in-house collaboration with the Macromolecular Crystallography Unit; or with Dr.Oliver Einsle from Freiburg University) or by solution-state NMR (in collaboration with Dr. Mario Piccioli and Dr.Francesca Cantini from CERM, Florence). Within the solution-state NMR, IBN is also involved in method development for taking full advantage of the paramagnetic properties that naturally occur in these metalloproteins.
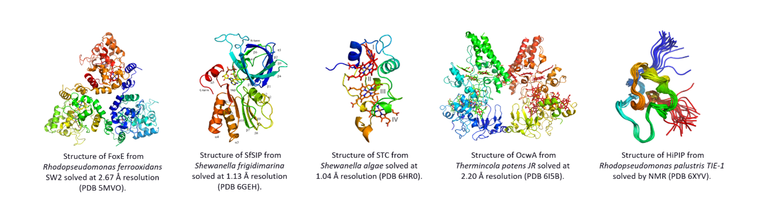
Relevant literature:
1. Trindade IB, Invernici M, Cantini F, Louro RO*, Piccioli M*, PRE‐driven Protein NMR Structures: an Alternative Approach in Highly Paramagnetic Systems, FEBS J, (2021) 288, 3010-3023 DOI:10.1111/febs.15615
2. Costa NL, Herman B, Fourmond V, Faustino MM, Teixeira M, Einsle O, Paquete C M, Louro RO, How thermophilic Gram-positive organisms perform extracellular electron transfer: characterization of the cell surface terminal reductase OcwA, mBio (2019) 10, e1210-19. DOI: 10.1128/mBio.01210-19
3. Fonseca BM, Silva L, Trindade IB, Moe E, Matias PM, Louro RO, Paquete CM, Optimizing electroactive organisms: the effect of orthologous proteins, Frontiers in Energy Research (2019) 7, DOI: 10.3389/fenrg.2019.00002
4. Trindade IB, Silva JM, Fonseca BM, Catarino T, Fujita M, Matias PM, Moe E, Louro RO Structure and reactivity of a siderophore-interacting protein from the marine bacterium Shewanella reveals unanticipated functional versatility, J Biol Chem (2019) 294, 157-167 DOI: 10.1074/jbc.RA118.005041
5. Pereira L, Saraiva IH, Oliveira ASF, Soares CM, Louro RO*, Frazão C*, Molecular structure of FoxE, the putative iron oxidase of Rhodobacter ferrooxidans SW2, Biochim Biophys Acta-Bioenergetics 1858, 847-853 (2017 )DOI: 10.1016/j.bbabio.2017.07.002



