Microbial & Enzyme Technology Lab
TOWARD A BIO-BASED SOCIETY
Industrial Biotechnology aims to use the power of enzymes and microorganisms to craft bioproducts, biomaterials, energy, and fuels from renewable sources. It offers environmentally friendly alternatives to traditional chemical methods.
Our laboratory is dedicated to exploring new bacterial enzymes involved in lignocellulose degradation through comprehensive biochemical and structural characterization and enhancing their properties using protein engineering tools. We additionally focus on developing eco-friendly and cost-effective bioprocesses, looking for green methodologies and synthesis of bioactive molecules.
Current work employs a combination of enzymology, molecular biology, structural biology, and microbiology, and the research is at the crossroads of protein science and technology. We aim to expand the range of biocatalysts and are committed to uncovering the intricate relationship between enzyme function and structure and delving into the molecular mechanisms behind enzyme evolution.
Curious about what we do? Watch this short video to explore METlab’s science, people, and impact!
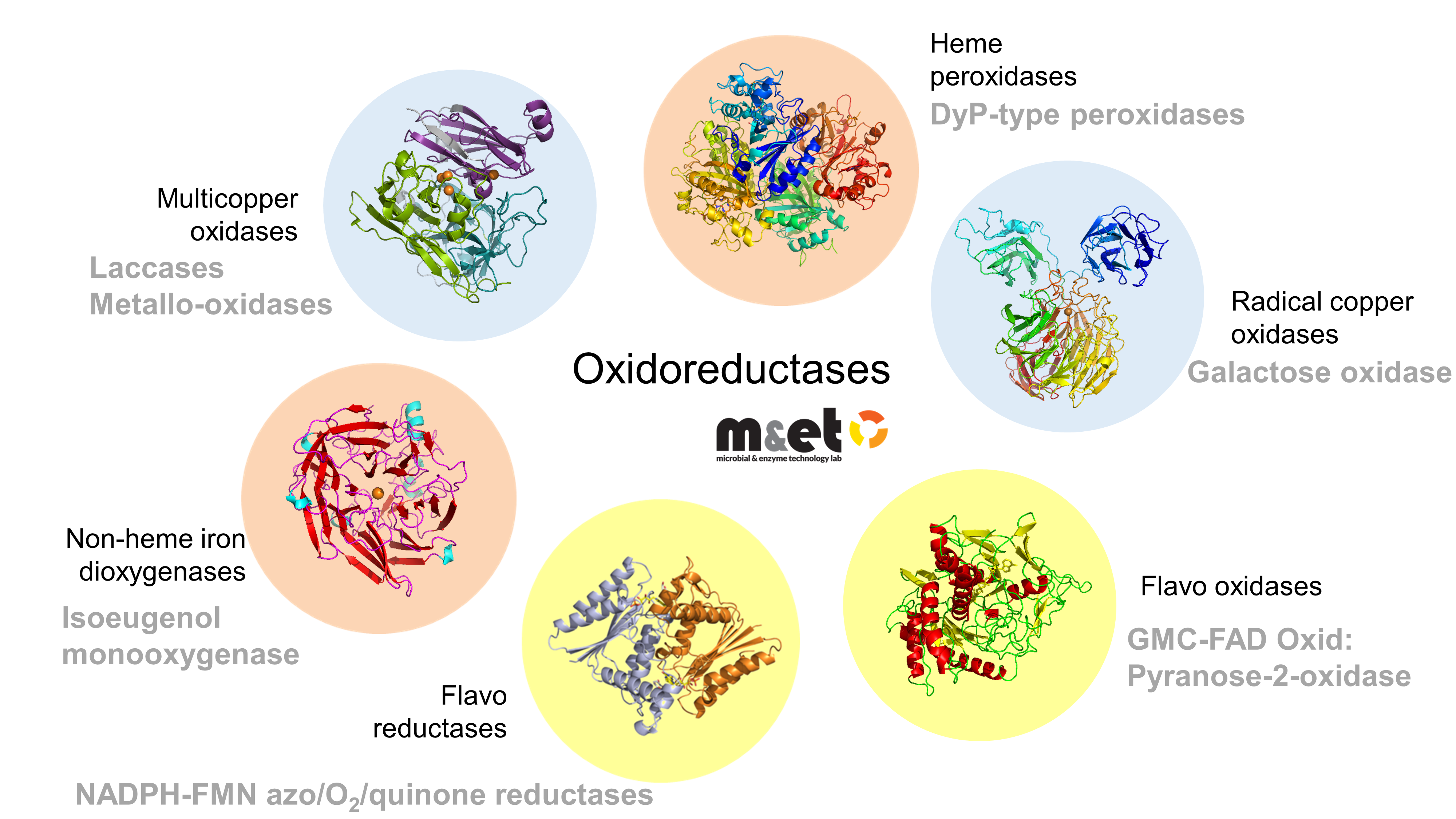
Enzyme Systems and Biocatalysis
Our group explores and engineers oxidoreductive enzymes for sustainable biotechnological applications. We combine structural biology, enzymology, and protein engineering to understand catalytic mechanisms and create robust biocatalysts for lignin valorization, green chemistry, and industrial bioprocesses.
Multicopper Oxidases (Laccases)
We pioneered the structural and functional characterization of bacterial laccases, key ligninolytic enzymes that oxidize a wide range of aromatic substrates using only oxygen. Our work revealed how electrostatics and copper assembly control activity and stability, and guided the development of engineered variants with tunable substrate specificity. These enzymes are being applied to valorize lignin-derived compounds for bio-based chemicals and materials.
Azoreductases and DyP-Peroxidases
We identified and optimized bacterial azoreductases and peroxidases capable of degrading synthetic dyes and transforming aromatic compounds into valuable building blocks. Using directed evolution, we enhanced enzyme stability and catalytic efficiency, paving the way for cost-effective and eco-friendly bioprocesses.
Pyranose and Galactose Oxidases
We discovered and characterized new bacterial sugar oxidases with unique activities and selectivities. Recent work on glycose 3-oxidase (PsG3Ox) and galactose oxidase variants has led to the efficient synthesis of rare sugars and aldehydes from renewable feedstocks — processes with strong potential for industrial-scale up and technology transfer.
Isoeugenol Dioxygenases
In the context of lignin valorization, we study enzymes that convert isoeugenol directly into vanillin. Through computational design and immobilization strategies, we have developed improved variants with high catalytic efficiency and stability, enabling greener production of bio-based aroma compounds.



