Moe Team || DNA repair
Highlights
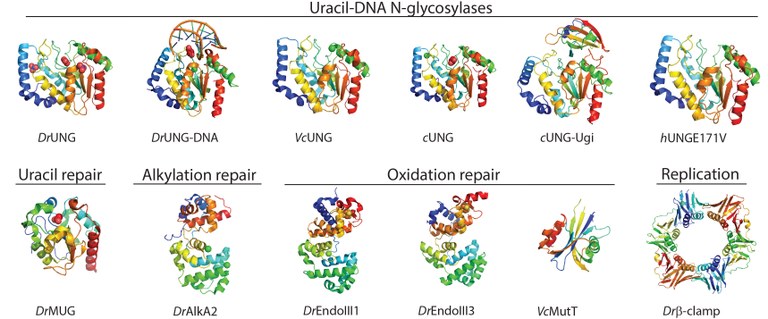
Team members
Elin Moe, PhD
Andreia Fernandes, PhD student
Filipe Rollo, PhD student (shared with Dr. Smilja Todorovic,
Carlota Conceição, PhD student (visitor from FCT)
Collaborations
Institut de Biologie Structurale (IBS), Grenoble, France.
Department of cancer research and molecular medicine, NTNU, Norway.
Department of Chemistry, UiT – the arctic university of Norway.
Egas Moniz – Cooperativa de Ensino Superior (CESEM) and UCIBIO@FCT UNL, Portugal.
Technische Universität Berlin, Germany.
Publications (2015 - 2022)
Moe E, Silveira CM, Zuccarello L, Rollo F, Stelter M, De Bonis S, Kulka-Peschke C, Katz S, Hildebrandt P, Zebger I, Timmins J, Todorovic S. Human endonuclease III/NTH1: focusing on the [4Fe-4S] cluster and the N-terminal domain. Chem Commun (Camb). 2022 Nov 10;58(90):12568-12571. [doi: 10.1039/d2cc03643f. PMID: 36279116].
Rollo F, Borges PT, Silveira CM, Rosa MTG, Todorovic S, Moe E. Disentangling Unusual Catalytic Properties and the Role of the [4Fe-4S] Cluster of Three Endonuclease III from the Extremophile D. radiodurans. Molecules. 2022 Jul 2;27(13):4270. doi: 10.3390/molecules27134270. PMID: 35807515; PMCID: PMC9268735.
Silveira, C. M., Moe, E., Fraaije, M., Martins, L. O., & Todorovic, S. (2020). Resonance Raman view of the active site architecture in bacterial DyP-type peroxidases. RSC Advances. https://doi.org/10.1039/d0ra00950d
Trindade, Inês B, Moe, E., & Louro, R. O. (2020). Siderophore‐Interacting Protein. In Encyclopedia of Inorganic and Bioinorganic Chemistry. https://doi.org/10.1002/9781119951438.eibc2743
Fonseca, B. M., Silva, L., Trindade, I. B., Moe, E., Matias, P. M., Louro, R. O., & Paquete, C. M. (2019). Optimizing Electroactive Organisms: The Effect of Orthologous Proteins. Frontiers in Energy Research, 7. https://doi.org/10.3389/fenrg.2019.00002
Trindade, I.B., Silva, J. M., Fonseca, B. M., Catarino, T., Fujita, M., Matias, P. M., Moe, E., & Louro, R. O. (2019). Structure and reactivity of a siderophore-interacting protein from the marine bacterium Shewanella reveals unanticipated functional versatility. Journal of Biological Chemistry, 294(1). https://doi.org/10.1074/jbc.RA118.005041
Sarre, A., Stelter, M., Rollo, F., De Bonis, S., Seck, A., Hognon, C., Ravanat, J.-L., Monari, A., Dehez, F., Moe, E., & Timmins, J. (2019). The three Endonuclease III variants of Deinococcus radiodurans possess distinct and complementary DNA repair activities. DNA Repair, 78. https://doi.org/10.1016/j.dnarep.2019.03.014
Frade, K. S. T., Fernandes, A. C. P., Silveira, C. M., Fraza-o, C., & Moe, E. (2018). A novel bacterial class v dye-decolourizing peroxidase from the extremophile Deinococcus radiodurans: Cloning, expression optimization, purification, crystallization, initial characterization and X-ray diffraction analysis. Acta Crystallographica Section F: Structural Biology Communications. https://doi.org/10.1107/S2053230X18008488
Moe, Elin, Rollo, F., Silveira, C. M., Sezer, M., Hildebrandt, P., & Todorovic, S. (2018). Spectroelectrochemical insights into structural and redox properties of immobilized endonuclease III and its catalytically inactive mutant. Spectrochimica Acta - Part A: Molecular and Biomolecular Spectroscopy, 188, 149–154. https://doi.org/10.1016/j.saa.2017.06.050
Timmins, J., & Moe, E. (2016). A decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: Major findings, functional and mechanistic insight and challenges. Computational and Structural Biotechnology Journal, 14, 168–176. https://doi.org/doi:10.1016/j.csbj.2016.04.001
Trindade, I.B., Fonseca, B. M., Matias, P. M., Louro, R. O., & Moe, E. (2016). A putative siderophore-interacting protein from the marine bacterium Shewanella frigidimarina NCIMB 400: Cloning, expression, purification, crystallization and X-ray diffraction analysis. Acta Crystallographica Section:F Structural Biology Communications, 72. https://doi.org/10.1107/S2053230X16011419
Niiranen, L., Lian, K., K., J., & Moe, E. (2015). Crystal structure determination of DNA polymerase III b-subunit from the extreme radiation and desiccation resistant bacterium Deinococcus radiodurans. BMC Structural Biology, 15:5. https://doi.org/DOI: 10.1186/s12900-015-0032-6
Lian, K., Leiros, H. K. S., & Moe, E. (2015). MutT from the fish pathogen Aliivibrio salmonicida is a cold-active nucleotide-pool sanitization enzyme with unexpectedly high thermostability. FEBS Open Bio, 5. https://doi.org/10.1016/j.fob.2015.01.006
Moe, E, Assefa, N. G., Leiros, I., Torseth, K. J., Smalås, A. O., & Willassen, N. P. (2015). Reduced Hydrophobicity of the Minor Groove Intercalation Loop is Critical for Efficient Catalysis by Cold Adapted Uracil-DNA N-Glycosylase from Atlantic Cod. J Thermodyn Catal, 6:155. https://doi.org/10.4172/2157-7544.1000155
Sarre, A., Ökvist, M., Klar, T., Hall, D. R., Smalås, A. O., McSweeney, S., Timmins, J., & Moe, E. (2015). Structural and functional characterization of two unusual endonuclease III enzymes from Deinococcus radiodurans. Journal of Structural Biology, 191(2). https://doi.org/10.1016/j.jsb.2015.05.009
Pedersen, H. L., Johnson, K. A., Mcvey, C. E., Leiros, I., & Moe, E. (2015). Structure determination of uracil-DNA N-glycosylase from Deinococcus radiodurans in complex with DNA. Acta Crystallographica Section D: Biological Crystallography, 71. https://doi.org/10.1107/S1399004715014157
Moe, E, Sezer, M., Hildebrandt, P., & Todorovic, S. (2015). Surface enhanced vibrational spectroscopic evidence for an alternative DNA-independent redox activation of endonuclease III. Chem Commun (Camb), 51(15), 3255–3257. https://doi.org/10.1039/c4cc09498k
