McVey Lab - Structural Virology
 The topic of virus–host cell interactions spans all of virology and provides some of the most important insights into this field including the pathological effects of these interactions, and therapeutic interventions.
The topic of virus–host cell interactions spans all of virology and provides some of the most important insights into this field including the pathological effects of these interactions, and therapeutic interventions.
We are studying the mechanisms involved in the establishment and modulation of herpesviral chromatin. Chromatin modulation is a key process that governs the infectious or persistent cycle of herpesviruses. The long-term goal is to understand how these processes contribute to virus tumourigenesis. To accomplish this, we investigate the following topics:
- The relationship between the viral protein LANA and its interaction with a plethora of host cell proteins.
- The abilities of viral macromolecules to interact and modulate host chromatin architecture, including those that impact human health.
The research performed by our laboratory requires an integrated approach wherein techniques such as protein interaction analysis use X-ray crystallography, BioSAXS, ITC and EMSA to understand the components of viral modulation and the processes that initiate and maintain viral latency.

Highlights
December 2021
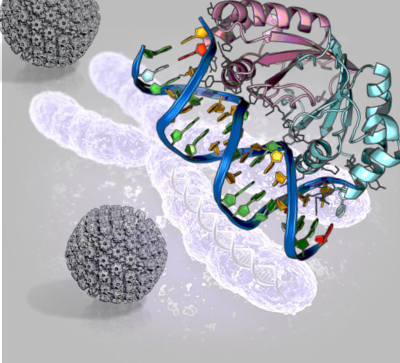
Kaposi’s sarcoma-associated herpesvirus (KSHV) is one of seven currently known human cancer viruses, or oncoviruses, and a leading cause of  malignancy in AIDS. KSHV establishes persistent, lifelong infection of its human host and persists latently in tumor cells. In a new study, researchers revealed how virus latency is linked to the Mixed-lineage leukemia 1 protein.
malignancy in AIDS. KSHV establishes persistent, lifelong infection of its human host and persists latently in tumor cells. In a new study, researchers revealed how virus latency is linked to the Mixed-lineage leukemia 1 protein.
During latency, gammaherpesviruses, such as KSHV, persist as multicopy, circularized genomes in the cell nucleus (episome) and express a small subset of viral genes. KSHV latency-associated nuclear antigen (LANA) is the predominant gene expressed. LANA, plays a critical role during latent infection and causes several KSHV associated malignancies. Upon infection, the viral genome is rapidly transported to the nucleus, where it associates with histone proteins to form chromatin, this compacts the viral DNA and plays a role in replication and life cycle of the virus. This process involves several interactions with host proteins, such as Mixed Lineage Leukemia 1 (MLL1), and leads to chromatin modifications. MLL1 is a histone methyltransferase, and when deregulated is related to pediatric and adult leukemia.
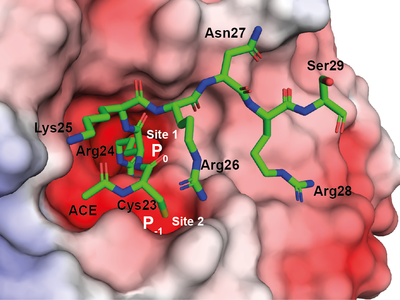
Through the elucidation of the X-ray crystal structure of a LANA peptide bound to WDR5, a component of the MLL1 methylase complex, revealed a potential regulatory mechanism. When the leukemia-related protein expression is disrupted, the capacity of KSHV to develop latency is extremely poor.
This discovery represents an important step in the effort for the prevention and treatment of KSHV malignancy. “By understanding in detail how the KSHV LANA protein and MLL1 interact, it is possible to develop strategies to disrupt MLL1 regulation and function of KSHV”.
Team members
Colin E. McVey, DPhil, Lab Head
Collaborations
- Marta Pires de Miranda (Católica Biomedical Research Centre, Lisbon, PT)
- Kenneth Kaye (Harvard Medical School, USA)
- Pedro Simas (Católica Biomedical Research Centre, Lisbon, PT)
- Dmitry Semchonok (Cryo-EM Integrative Structural Biology Laboratory, ITQB NOVA, Oeiras, PT)
- Tiago N. Cordeiro (Dynamic Structural Biology Lab, ITQB NOVA, Oeiras, PT)
Selected Publications
-
Tan, M., Li, S., Juillard, F., Chitas, R., Custodio, T.F., Xue, H., Szymula, A., Sun, Q., Liu, B., Alvarez, A.L., Chen, S., Huang, J., Simas, J.P., McVey, C.E., Kaye, K.M., 2021. MLL1 is regulated by KSHV LANA and is important for virus latency. Nucleic acids research 49, 12895–12911. https://doi.org/10.1093/nar/gkab1094
-
Pires de Miranda, M., Quendera, A.P., McVey, C.E., Kaye, K.M., Simas, J.P., 2018. In Vivo Persistence of Chimeric Virus after Substitution of the Kaposi’s Sarcoma-Associated Herpesvirus LANA DNA Binding Domain with That of Murid Herpesvirus 4. J Virol 92. https://doi.org/10.1128/JVI.01251-18
-
Habison, A.C., de Miranda, M.P., Beauchemin, C., Tan, M., Cerqueira, S.A., Correia, B., Ponnusamy, R., Usherwood, E.J., McVey, C.E., Simas, J.P., Kaye, K.M., (2017). Cross-species conservation of episome maintenance provides a basis for in vivo investigation of Kaposi’s sarcoma herpesvirus LANA. PLoS pathogens 13, e1006555. https://doi.org/10.1371/journal.ppat.1006555
-
S.A. Cerqueira, M.Tan, S. Li, F. Juillard, C.E. McVey, K.M. Kaye, J.P. Simas (2016) "LANA E3 Ubiquitin-Ligase Activity Impacts Gammaherpesvirus-Driven Germinal Center B Cell Proliferation", J Virol. 2016 Jun 15.
http://dx.doi.org/10.1128/JVI.00813-16 -
Ponnusamy, M.V. Petoukhov, B. Correia, T.F. Custodio, F. Juillard, M. Tan, M. Pires de Miranda, M.A. Carrondo, J.P. Simas, K.M. Kaye, D.I. Svergun, C.E. McVey (2015) "KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA." Nucleic Acids Res. 43 (20): 10039-54. http://dx.doi.org/10.1093/nar/gkv987
- Correia, S.A. Cerqueira, C. Beauchemin, M. Pires de Miranda, S. Li, R. Ponnusamy, L. Rodrigues, T. R. Schneider, M. A. Carrondo, K. M. Kaye, J. P. Simas and C. E. McVey “Crystal structure of the gamma-2 herpesvirus LANA DNA binding domain identifies charged surface residues which impact viral latency", PLoS Pathog. (2013) 9(10): e1003673. doi:10.1371/journal.ppat.1003673.
