Doing things differently
Oeiras, 03.10.11
An international team that includes researchers from the Lab of Molecular Genetics of Microbial Resistancehas unraveled a new biosynthetic pathway of the heme group, which is formed by an atom of iron contained in the center of an organic cyclic compound named porphyrin, and is required for the function of several essential proteins. Results were gathered from three different types of ancient organisms: archaea, denitrifying bacteria and sulfate reducing bacteria. The findings are published in the current issue of PNAS.
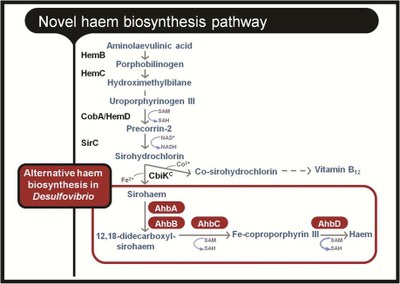
ITQB researchers concentrated on heme biosynthesis in the sulfate reducing bacteria Desulfovibrio. How the heme is formed in Desulfovibrio has been a mystery. The typical biosynthetic pathway was known in other organisms but the Desulfovibrio genome lacked the corresponding genes (and thus the enzymes). This set ITQB researchers to search for candidates for the heme’s biosynthetic enzymes and genes. The biosynthetic pathway was reconstructed step by step in vitro leading researchers to confirm that in Desulfovibrio things take a slightly different route. At a certain point of the classical pathway, the siroheme intermediary is formed and a new branch is now used for synthesizing heme.
These results confirmed the researchers’ earlier proposal of an alternative pathway for heme biosynthesis in Desulfovibrio. Interestingly, and as shown in this paper, this pathway is also active in archaea and is probably used by denitrifying bacteria to synthesize a different kind of heme (heme d1), therefore suggesting an evolutionary precedence of this mechanism for heme production in living organisms.
Original Article
PNAS October 3, 2011 doi: 10.1073/pnas.1108228108
Molecular hijacking of siroheme for the synthesis of heme and d1 heme
Shilpa Bali, Andrew D. Lawrence, Susana A. Lobo, Lígia M. Saraiva, Bernard T. Golding, David J. Palmer, Mark J. Howard, Stuart J. Ferguson and Martin J. Warren







