Old bacteria with fancy tricks
Oeiras, 06.04.2015
Even in organisms as simple as bacteria, cell differentiation requires a precise temporal and spatial regulation of gene expression. Researchers from the Microbial Developmental Lab and collaborators found a sophisticated genetic regulatory mechanism in bacteria, resembling those found in higher organisms. The work is published in PLOS Genetics.
Some bacteria, like Bacillus subtilis, exposed to adverse environmental conditions are able to differentiate a highly resistant type of cell – the spore. The process of spore formation involves only two cell types – the forespore, which will give rise to the spore, and the mother cell, inside which the spore is formed – and as such it is a great model for cell differentiation. During this process, which takes 6 to 8 hours in the lab, more than 10% of the genome is recruited. This means hundreds of genes that have to be expressed in the right cell at the right time. To make sure spore formation is done properly nothing can be left to chance.
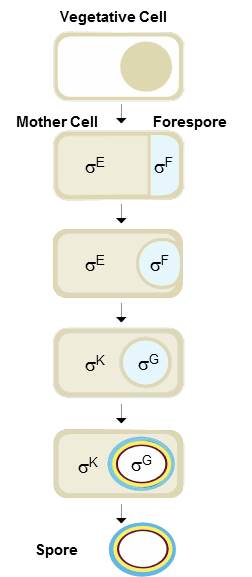 The formation of bacterial spores is controlled by specific proteins – the sigma factors - that act as switches, activating and repressing dozens of genes. There are four different sigma factors - F, G, K, and E – with a very precise action in time and space. In the spore, it is first sigmaF and then sigmaG; in the mother cell, it is first sigmaK and then sigmaE.
The formation of bacterial spores is controlled by specific proteins – the sigma factors - that act as switches, activating and repressing dozens of genes. There are four different sigma factors - F, G, K, and E – with a very precise action in time and space. In the spore, it is first sigmaF and then sigmaG; in the mother cell, it is first sigmaK and then sigmaE.
Researchers knew that the concerted action of sigma factors is controlled through a complex genetic mechanism that regulates the expression of sporulation genes. But they discovered an additional control mechanism at the level of the sigma factors themselves. This new mechanism involves a fifth protein, antisigma, a kind of main switch that ensures that sigma factor are only active when and where they are necessary.
The sophisticated mechanism of antisigma is different in the two bacterial cells. In the mother cell, antigma is produced under the control of the late sigmaK and blocks the action of the early sigmaE. As a result, all genes controlled by sigmaE are switched off and sigmaK works alone in the final stages of development. On the other hand, in the forespore, antisgma is produced under the control of the early sigmaF and binds to the late sigma G, blocking its action in the early stages of development. This complex mechanism surprised the scientists: “when we look at the molecular details, the sophistication is even more incredible, especially if we think that the ability to form spores appeared 2,8 billion years ago”, says Mónica Serrano, first author of the study.
Original Article
PLOS Genetics (2015) DOI: 10.1371/journal.pgen.1005104
Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in Bacillus subtilis
Mónica Serrano, JinXin Gao, João Bota, Ashley R. Bate, Jeffrey Meisner, Patrick Eichenberger, Charles P. Moran Jr., Adriano O. Henriques
Mónica Serrano, João Bota, Adriano O. Henriques - ITQB
JinXin Gao, Jeffrey Meisner, Charles P. Moran Jr. - Emory University School of Medicine, (USA)
Ashley R. Bate, Patrick Eichenberger - New York University (USA)







