Research Highlights
 |
|||
ITQB News
A recent collaboration protocol between NOVA and Pfizer will be reinforcing the areas of research and development, as well as education and literacy
Oeiras, 18.03.2021
|
Universidade NOVA de Lisboa and the biopharmaceutical company Pfizer have signed, at the end of January 2021, a collaboration protocol in the areas of Research & Development, Postgraduate Education, and Education for Health and Literacy. Aware of the relevance and impact of chronic diseases and infection, the Chronic Disease and Infection group, one of NOVA health's strategic areas, promoted this link between NOVA and Pfizer |
 |
This group, co-coordinated by researcher Lígia Saraiva at ITQB NOVA, also promotes "Think Tanks" with the goal of developing cooperation mechanisms between the two entities.
With this partnership, NOVA reinforces its mission towards collaborative, responsible, and internationally relevant research, which prioritizes the resolution of problems that affect the population, namely in the area of chronic diseases and infection.
Mechanism of crosstalk between systems for haem biosynthesis and external capture in pathogens published today
Oeiras, 10.07.2018
We describe how the human pathogen Staphylococcus aureus deals with this conundrum. The protein-protein interaction that occurs between IsdG, one of two S. aureus haem monooxygenase enzymes that belong to the external haem uptake system, and ferrochelatase, the penultimate enzyme of the S. aureus haem biosynthetic pathway, inhibits the endogenous formation of haem. This seems to be a general mechanism as IsdG homologues appear widespread in many pathogens, and we propose that this crosstalk reconciles their high haem requirements with the paradox that excess of haem is lethal”, according to Lígia Saraiva, corresponding author.
This work was developed with colleagues from Instituto Superior Técnico (Portugal), University of Kent (UK) and University of Vienna (Austria).

Original Paper
Molecular Microbiology 2018 doi.org/10.1111/mmi.14060
Staphylococcus aureus haem biosynthesis and acquisition pathways are linked through haem monooxygenase IsdG
Marco A.M. Videira, Susana A.L. Lobo, Liliana S.O. Silva, David J. Palmer, Martin J. Warren, Manuel Prieto, Ana Coutinho, Filipa L. Sousa, Fábio Fernandes and Lígia M. Saraiva*
Evolution of gene duplication in Trichomonas vaginalis
[Survival strategies for microorganisms]
Oeiras, 29.06.2016
|
Trichomoniasis is a sexually transmitted disease (STD) caused by the parasiteTrichomonas vaginalis. Trichomoniasis is the most common curable STD in young, sexually active women. According to the WHO, an estimated 160 million new cases occur each year in women and men, although men often do not have any symptoms and can be undiagnosed carriers. Pathogens suffer damages inflicted by the chemicals that are produced by the host immune system during the infectious process. However, pathogens evolve several means of evading or subverting the host defences. ITQB NOVA’s Molecular Mechanisms of Pathogen Resistance Laboratory previously revealed a new family of protein, the Repair of Iron Centres proteins (RICs) that protects microbes from the damage caused on pathogens by the oxidative and nitrosative stress imposed by the host, and showed that RICs are widely spread in nature from bacteria, fungi and protozoa. |
|
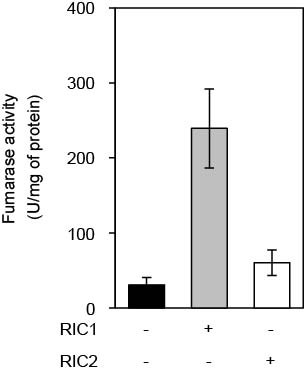
Interestingly, T. vaginalis is the only microbe that expresses two homologs, named RIC1 and RIC2. In this study the authors have addressed this gene duplication phenomenon. It is shown that the two T. vaginalis RICs share significant amino acid sequence similarity but have different biochemical characteristics that provide different functions.
This paper has been recommended by David Leitsh, University of Vienna, on F1000 Prime, an in-depth directory of the top articles in biology and medicine. Articles are selected by a peer-nominated global 'Faculty' of the world's leading scientists and clinicians who then rate them and explain their importance.
“This paper constitutes the first study on 'repair of iron centres' proteins (RICs) of a protist parasite, i.e. of Trichomonas vaginalis. In fact, RICs have not been identified in any organism outside the bacterial kingdom so far, rendering this study all the more interesting.”
in Leitsch D: F1000Prime Recommendation of [Nobre LS et al., Protist 2016, 167(3):222-233]. In F1000Prime, 08 Jun 2016; DOI: 10.3410/f.726321114.793519156.
Original article
Protist, Vol. 167, 222–233, June 2016
Trichomonas vaginalis Repair of Iron Centres Proteins: The Different Role of Two Paralogs
Lígia S. Nobre, Dionigia Meloni, Miguel Teixeira, Eric Viscogliosi, and Lígia M. Saraiva
Double effect against inflammation: CO releasing molecule works best when coupled to anti-inflammatory drug
Oeiras, 04.09.2015
| Carbon monoxide is also known for its anti-inflammatory properties and different strategies are being sought for delivering it to the organism. A team of researchers involving the company Alfama, the Lab of Organometallic Chemistry, and the Lab of Molecular Genetics of Microbial Resistance, together with a team from iMMLisboa led by Gonçalo Bernardes, has developeda new carbon monoxide releasing molecule coupled with a drug with anti-oxidant and anti-inflammatory properties, which elicits an enhanced anti-inflammatory effect in vitro. The study is published as a HOT paper in Chemistry - A European Journal. | 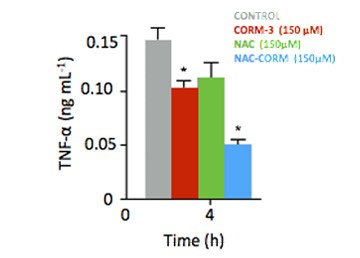 |
“What we found, even if these are early findings, is that by combining the anti-inflammatory drug N-Acetylcysteine (NAC) with a ruthenium compound able to release carbon monoxide, we achieve a double anti-inflammatory effect” explains João Seixas first author of the paper formerly at Alfama and ITQB, presently at iMMLisboa. “This compound is able to both deliver biologically active carbon monoxide and to scavenge the reactive oxygen species, known to be formed during the carbon monoxide release process and which contribute to the inflammation response”. The data suggests that similar compounds may also act synergistically to elicit an enhanced anti-inflammatory response.
“This work really strengthens previous work in carbon monoxide therapy, an area we have led in the last decade” says Carlos Romão, head of the Lab of Organometallic Chemistry and co-founder of Alfama, a pharmaceutical company focusing on carbon monoxide releasing molecules.
The paper has also the collaboration from researchers from University of Cambridge (UK) and the University of Cartagena (Colombia).
Original Article
Chemistry - A European Journal (2015) Early View DOI: 10.1002/chem.201502474
An N-Acetyl Cysteine Ruthenium Tricarbonyl Conjugate Enables Simultaneous Release of CO and Ablation of Reactive Oxygen Species
João D. Seixas1,2,3, Miguel Chaves-Ferreira1, Diana Montes-Grajales 4,5, Ana M. Gonçalves3, Ana R. Marques3, Lígia M. Saraiva2, Jesus Olivero-Verbel5, Carlos C. Romão2,3 and Gonçalo J. L. Bernardes1,4
1 - iMM Lisboa, 2- ITQB, 3 - Alfama, 4 - University of Cambridge (UK), 5 - University of Cartagena (Colombia)
Odd enzymes as antibiotic targets: Researchers explore unconventional haem biosynthesis in Staphylococcus aureus
Oeiras, 04.05.2015
| The haem molecule is found in all kingdoms of life, for example in humans is an important part of haemoglobin, but not all organisms obtain it the same way. Researchers from the Molecular Genetics of Microbial Resistance Lab and collaborators demonstrated that haem biosynthesis in the bacteriaStaphylococcus aureus follows an unconventional path, and explored the potential of these biochemical differences as targets for novel antimicrobial agents. The work is published in Molecular Microbiology. | 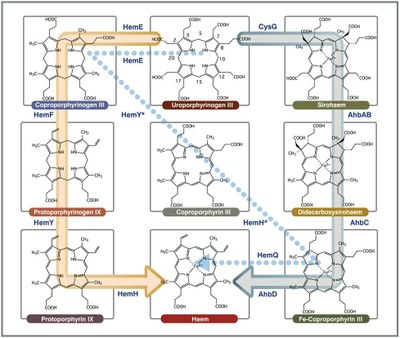 |
Staphylococcus aureus are pathogenic bacteria widely known for their increasing resistance to commonly used antibiotics therefore all singularities are worth exploring as potential weaknesses. The haem molecule isessential for these bacteria, which are able both to acquired it from the environment and synthesizing it from scratch. In the work now reported, researchers fully characterized the enzymes involved in haem biosynthesis in Staphylococcus and concluded that the biochemical pathway, named transitional pathway, lies somewhere in between the more widely known pathway and the alternative pathway active in ancient bacteria, also recently found by the authors of this study.
Staphylococcus uses enzymes different from humans, so these become possible targets for new antibiotics. To check if the biochemical differences for haem biosynthesis could be explored to fight Staphylococcus aureus, researchers screened 52 different compounds as potential inhibitors of one of the enzymes (HemY). Several compounds were shown to inhibit HemY and thus have the potential for further development. Because the transitional pathway for haem production identified in Staph is restricted to many Gram-positive bacteria, researchers propose that these enzymes may be used for selective therapy to differentiate between Gram-positive and Gram-negative pathogens.
Original Article
Molecular Microbiology (2015) Accepted Article DOI: 10.1111/mmi.13041
Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway
Susana AL Lobo1,4, Alan Scott2,4, Marco AM Videira1, David Winpenny3, Mark Gardner3, Mike J Palmer3, Susanne Schroeder2, Andrew Lawrence2, Tanya Parkinson3, Martin Warren2 and Lígia M Saraiva1
1 - Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras, Portugal
2 - University of Kent, UK
3 - Pfizer Global Research and Development, UK
4 - Both authors contributed equally to this work and should be considered co-first authors
Novel bacterial strategy to cope with host defenses uncovered in Helicobacter pylori: an unprecedented nitric oxide detoxifying system
Oeiras, 01.02.12
| Pathologies like chronic gastritis and gastric ulcers are caused by a bacterium, Helicobacter pylori, able to bypass the immunity control systems and to sustain long term colonization inside the harsh environment of the stomach. Now, ITQB researchers from the Molecular Genetics of Microbial Resistance Lab and collaborators from the Institut Pasteur in Paris have identified a new protein in Helicobacter pylori, which assures the bacterial survival inside the host by protecting bacteria against the effects of nitric oxide. The work is published in the journal Antioxidants and Redox Signaling. |
|
|
Many bacteria resort to nitric oxide (NO) detoxification systems to counteract the host NO production, a crucial tool of mammal’s innate immunity for the control and clearance of pathogens. The Gram negative bacteria H. pylori lack the typical bacterial systems, which suggested to researchers that other unknown proteins could be involved in NO detoxification. After screening a collection of mutants for unusual growth in the presence of NO, researchers identified a gene, unknown till then, that confers resistance to NO.
The next step was to gather clues about this new gene’s function, resorting to genetics, biochemistry and molecular biology. Clue number 1, the mutant H. pylori was more susceptible to nitric oxide than the wild type strains. Clue number 2, mutants were also less able to survive upon incubation with the immune system macrophages. Clue number 3, mutants were less efficient in colonizing the mice themselves. Clue number 4,if mice lacked the NO production mechanism, mutant bacteria were as efficient as wild type. Clue number 5, the purified wild type protein was an active NO reductase. Clue number 6, sequence analysis revealed that the new gene is present in several other prokaryotes. Conclusion, a previously hypothetical protein of H. pylory is now renamed NorH, for NO reductase of Helycobacter pylori, and represents a new family of enzymes for microbial protection against nitric oxide associated stress.This study was jointly coordinated by Lígia Saraiva, head of the Molecular Genetics of Microbial Resistance Lab, and Ivo Boneca, ITQB alumnus now at the Institut Pasteur.
Original Article
Antioxidants & Redox Signaling, ahead of print. doi:10.1089/ars.2011.4304.
Helicobacter pylori has an unprecedented nitric oxide detoxifying system
Marta C. Justino1,2, Chantal Ecobichon2,3, André F. Fernandes1, Ivo G. Boneca2,3 and Lígia M. Saraiva1
1 - Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras, Portugal
2 - Institut Pasteur, Group Biology and Genetics of the Bacterial Cell Wall, Paris, France
3 - INSERM, Groupe AVENIR, Paris, France
Doing things differently: an alternative route for heme production unraveled in ancient organisms
Oeiras, 03.10.11
 |
An international team that includes researchers from the Lab of Molecular Genetics of Microbial Resistancehas unraveled a new biosynthetic pathway of the heme group, which is formed by an atom of iron contained in the center of an organic cyclic compound named porphyrin, and is required for the function of several essential proteins. Results were gathered from three different types of ancient organisms: archaea, denitrifying bacteria and sulfate reducing bacteria. The findings are published in the current issue of PNAS. |
|
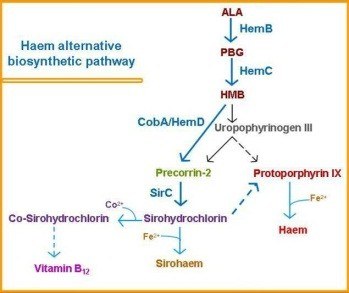
ITQB researchers concentrated on heme biosynthesis in the sulfate reducing bacteria Desulfovibrio. How the heme is formed in Desulfovibrio has been a mystery. The typical biosynthetic pathway was known in other organisms but the Desulfovibrio genome lacked the corresponding genes (and thus the enzymes). This set ITQB researchers to search for candidates for the heme’s biosynthetic enzymes and genes. The biosynthetic pathway was reconstructed step by step in vitro leading researchers to confirm that in Desulfovibrio things take a slightly different route. At a certain point of the classical pathway, the siroheme intermediary is formed and a new branch is now used for synthesizing heme. |
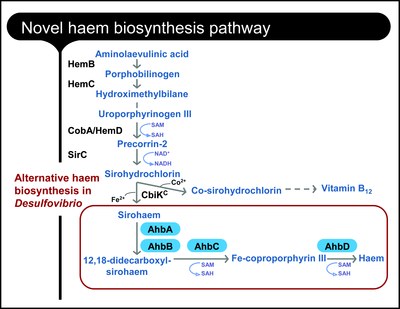 |
These results confirmed the researchers’ earlier proposal of an alternative pathway for heme biosynthesis in Desulfovibrio. Interestingly, and as shown in this paper, this pathway is also active in archaea and is probably used by denitrifying bacteria to synthesize a different kind of heme (heme d1), therefore suggesting an evolutionary precedence of this mechanism for heme production in living organisms.
Original Article
PNAS October 3, 2011 doi: 10.1073/pnas.1108228108
Molecular hijacking of siroheme for the synthesis of heme and d1 heme
Shilpa Bali, Andrew D. Lawrence, Susana A.L. Lobo, Lígia M. Saraiva, Bernard T. Golding, David J. Palmer, Mark J. Howard, Stuart J. Ferguson and Martin J. Warren
Enzyme evolution pictured by X-ray crystallography: : Researchers solve structures of three different chelatases
[Enzyme evolution pictured by X-ray crystallography]
Oeiras, 05.01.11
| Blood owes its red colour to the essential hemoglobin cofactor, heme. Present in many other important proteins, hemes require the insertion of a specific metal ion in a ring of carbons and nitrogen atoms (porphyrin), a reaction performed by chelatase enzymes. ITQB researchers joined efforts with researchers from the University of Kent and Queen Mary University of London to reveal the molecular mechanism of cobaltochelatases. The article, now published in the Proceedings of the National Academy of Science USA, gives a beautiful evolutionary picture of the chelatase family. | 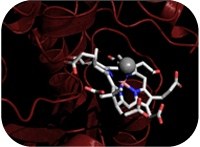 |
The simplest of cobaltochelatases is a small protein of two subunits present in archaea, a separate group of single-cell microorganisms. In most other organisms, the cobaltochelatases are about twice as big, a result of a gene duplication and fusion event. Resorting to X-ray crystallography, researchers solved the structure of chelatases from three different organisms - Archaeoglobus fulgidus (archaea), Salmonella enterica (Gram-positive bacteria), and Desulfovibrio vulgaris (Gram-negative bacteria). Because A. fulgidus and S. entericachelatases were caught with the porphyrin substrate and D. vulgaris chelatase with the cobalt ion, these structures provided molecular detail on the rearrangements that take place during the process of porphyrin chelation and insertion of cobalt.
ITQB researchers from the Macromolecular Crystallography Unit and the Molecular Genetics of Microbial Resistance Lab concentrated on the cobaltochelatase (CbiKP) from D. vulgaris, the more complex of these enzymes. A. fulgidus chelatase receives the substrate between two subunits; in S. enterica, these subunits maintain their structural identity but are fused in a single protein, the enzyme is actually a dimer with two matching active sites; D. vulgaris chelatase is a like a duplication of this dimer with two additional heme groups. Researchers found that, besides the four catalytic domains, the tetramer is organized in such a way that a central cavity with the potential for ligand binding is formed. Authors suggest that the cobaltochelatase (CbiKP) of D. vulgaris may have evolved an additional function, such as the transport of metals across the periplasmic space.
Original Article
Evolution in a family of chelatases facilitated by the introduction of active site asymmetry and protein oligomerization doi: 10.1073/pnas.1014298108 PNAS December 20, 2010
Célia V. Romão, Dimitrios Ladakis, Susana A. L. Lobo, Maria A. Carrondo, Amanda A. Brindley, Evelyne Deery, Pedro M. Matias, Richard W. Pickersgill, Lígia M. Saraiva, and Martin J. Warren
Killer or defender of S. aureus: antagonistic behaviour of a single enzyme. Study of a bifunctional enzyme with nitroreductase and GSNO reductase activity
Oeiras, 15.05.2009
| Nitrofuran derivatives are commonly used as antibiotics but require the presence of specific enzymes – nitroreductases - inside the bacteria to become active bactericides. When looking into the bacterial nitroreductases of Staphylococcus aureus, ITQB researchers encountered a novel nitroreductase that also has the ability to decompose S-nitrosoglutathione (GSNO), a compound formed in the bacteria upon exposure to the nitrosative stress generated by the immune system. The results are presented in the May issue of the Journal of Bacteriology. | 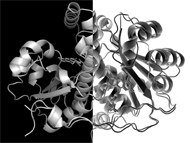 |
Staphylococcus aureus, a Gram-positive bacterium widely known by its increasing resistance to antibiotics, has four nitroreductases. By sequence analysis, researchers demonstrated that one of these nitroreductases belongs to a different subfamily and this spiked the interest in S. aureus nitroreductase NtrA.
Microorganisms exposed to nitrosative stress must deal with a variety of biochemical distresses, such as the damaging effect of GSNO, a product of intracellular glutathione’s reaction with NO. The effect of GSNO on essential bacterial proteins (through S-nitrosylation) is counteracted by its removal via GSNO reductase enzymes. Typically, these enzymes have additional functions in the cell but an enzyme that presented both GSNO reductase and nitroreductase activity had not yet been described.
Nitroreductase NtrA seems thus to play two opposite roles in Staphylococcus aureus; it promotes the activation of nitrofurans, which then become bactericidal compounds, and, in the absence of these antibiotics, it acts as a protection system against the harmful transnitrosylation reactions caused by endogenously formed GSNO.
Original paper
Journal of Bacteriology 191 (10): 3403-3406
A Novel Nitroreductase of Staphylococcus aureus with S-Nitrosoglutathione Reductase Activity
Ana Filipa N. Tavares, Lígia S. Nobre, Ana M. P. Melo, and Lígia M. Saraiva
Haem biosynthesis in anaerobic organisms: discovery of a novel haem alternative pathway
Oeiras, 23.04.09
| Haem cofactors are essential for the function of key proteins involved in a large array of metabolic processes. However, the enzymes involved in their biosynthesis remain uncharacterized in anaerobic organisms. To fill this gap the Molecular Genetics of Microbial Resistance Laboratory (and co-workers) recently characterized several of these proteins and the data provided insights for an alternative haem biosynthetic pathway present in some anaerobic bacteria and archaea. The results are published in the March issue of the Biochemical Journal.The authors suggest that the production of the haem group in primitive anaerobic enviroments probably required a different set of enzymes and that only later, as molecular oxygen became more abundant, the pathway involving several oxidative steps in the synthesis of haems became the more common route. | 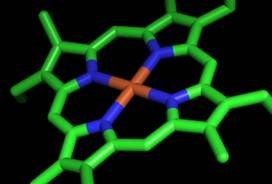 |
Original paper
Biochem. J. (2009) Immediate Publication, doi:10.1042/BJ20090151
Functional characterization of the early steps of tetrapyrrole biosynthesis and modification in Desulfovibrio vulgaris Hildenborough
Susana A. L. Lobo, Amanda Brindley, Martin J. Warren and Lígia M. Saraiva
| The article reports the comprehensive analysis of the haem biosynthetic pathway in the environmentally important anaerobic organism D. vulgaris Hildenborough. Several recombinant enzymes required for the transformation of 5-aminolevulinic acid into sirohydrochlorin and vitamin B12, were studied using molecular genetics and biochemistry methodologies. We show that porphobilinogen synthase (HemB) is a homohexamer zinc-dependent enzyme, and that porphobilinogen deaminase (HemC) contains a dipyrromethane cofactor. Interestingly, uroporphyrinogen III synthase is found fused with an uroporphyrinogen III methyltransferase (HemD-CobA) and its function was addressed by analysing not only the whole protein but also by dissecting the gene and studying the role of the individual protein domains. This study allowed to correct the genome annotation of a putative bifunctional precorrin-2 dehydrogenase/sirohydrochlorin ferrochelatase which was in fact shown to act only as a dehydrogenase and is simply capable of synthesizing sirohydrochlorin rather than sirohaem. |
|
In summary, the research described in this paper outlines how the basic tetrapyrrole framework is synthesized up to the first branch point, which is proposed to be sirohydrochlorin. The identification of bifunctional CobA/HemD is consistent with uroporphyrinogen III not being a branch point in the bifurcated pathway and represents a useful marker for an alternative haem biosynthetic route, which has been previously hypothesized to occur in some Desulfovibrio species and archaea.
Back to ITQB MGMR external page
News
Um prémio Nobel para como gerir a falta de oxigénio no nosso organismo
07.10.2019
A vida tal como a conhecemos não pode existir sem oxigénio. Contudo, as nossas células nem sempre dispõem de uma quantidade constante do elemento. Estados de hipoxia ocorrem em situações como o esforço físico ou as altitudes elevadas. A hipoxia resultante da diminuição do fluxo de sangue é também uma característica de doenças como a insuficiência cardíaca, doenças inflamatórias crónicas e vários tipos de cancro. O prémio Nobel da Medicina deste ano reconhece por isso a importância do trabalho de três cientistas que identificaram maquinarias celulares tais como o factor induzido por hipoxia – hypoxia-inducible factor HIF, que permitem ao nosso corpo detectar e adaptar-se às flutuações na quantidade de oxigénio a que as nossas células estão sujeitas – um grande passo em frente no conhecimento da fisiologia humana.
Os estudos agora distinguidos já levaram ao desenvolvimento (mas não ainda à utilização) de fármacos que interferem com a regulação do HIF e permitem a estimulação da produção de glóbulos vermelhos, ou de medicamentos que impedem a formação de novos vasos sanguíneos que facilitam o crescimento dos tumores. Também em Portugal, vários cientistas dedicam-se à investigação dos mecanismos de hipoxia e à sua associação a várias condições fisiológicas.
O oxigénio é ainda essencial a todos os microrganismos que produzem energia através de respiração celular. Uma diminuição excessiva da quantidade disponível para as células leva à sua morte. Assim, tal como os organismos superiores que respiram oxigénio dispõem de sistemas que permitem regular a quantidade acessível às células, e onde se enquadram os trabalhos sobre sensores de oxigénio dos laureados (neste caso, uma proteína contendo ferro), também as bactérias têm sistemas dedicados à detecção/resposta deste elemento. Sendo a microbiota parte essencial no funcionamento do nosso organismo, a sobrevivência dos microorganismos que a constituem está dependente da sua capacidade de adaptação às flutuações de oxigénio a que estão sujeitos.
O oxigénio tem ainda outra faceta: por reacção com componentes celulares, gera espécies muito mais reactivas que o próprio oxigénio, que podem levar à degradação de múltiplos processos metabólicos. Todos os organismos vivos, em particular os organismos anaeróbicos (que não necessitam de oxigénio para crescer), dispõem de sistemas para eliminar estas espécies reactivas de oxigénio. Estas espécies são utilizadas pelo sistema imunitário humano para combater microorganismos patogénicos.
No ITQB NOVA, investigamos a relação dos organismos com o oxigénio, nomeadamente em microorganismos unicelulares para os quais temos vindo a descobrir os mecanismos de defesa contra o oxigénio e as espécies reactivas derivadas. O seu estudo ao nível molecular através da determinação da sua estrutura, mecanismos de acção e contribuição relativa para a protecção das bactérias, é uma informação crucial para o conhecimento da interacção entre as células humanas e os microorganismos que competem pelas mesmas fontes de oxigénio. Ajuda também o desenvolvimento de novas estratégias que diminuam a sua resistência de bactérias patogenias. Esta simbiose entre microrganismos e células humanas é essencial e reflecte-se em semelhanças no seu modo de funcionamento: tal com as células animais contêm o regulador IHF para o oxigénio, as bactérias têm sensores de oxigénio (FNR, OxyR, …) que regulam a expressão de sistemas de resposta à sua deficiência ou excesso. No Ano Internacional da Tabela Periódica, este prémio Nobel sublinha a relevância de um dos mais importantes elementos químicos presentes na Terra, sem o qual parece que não sabemos (ainda) viver...

Grupo de Investigação estuda estratégias para combater a resistência a antibióticos
25.04.2019

Investigação em bactéria resistentes a antibióticos
28.12.2007

Saúde - Monóxido de carbono mata bactérias
12.2007
Investigação portuguesa conclui que monóxido carbono pode vir a ser usado como antibiótico
22.11.2007
Um estudo coordenado pela investigadora da Universidade Nova de Lisboa Lígia Saraiva conclui que o monóxido de carbono pode vir a ser usado como antibiótico, pela sua capacidade de matar bactérias, inclusive as mais resistentes.
"Não podemos prever quando é que teremos um antibiótico, o processo pode demorar muitos anos", afirmou a investigadora do Instituto de Tecnologia Química e Biológica (ITQB) da Universidade Nova de Lisboa em declarações à agência Lusa, sublinhando que os avanços conseguidos com a investigação representam apenas, por enquanto, um novo leque de possibilidades.
De acordo com o comunicado relativo à investigação, esta mostrou "pela primeira vez que o monóxido de carbono tem a capacidade de matar bactérias", algo demonstrado em "várias bactérias, em particular no bem conhecido patogénico Estafilococos, que tem vindo a desenvolver uma resistência preocupante aos antibióticos correntes".
"Os compostos libertadores de monóxido de carbono - CO, no símbolo químico - penetram as paredes celulares e só quando já estão dentro do meio celular, libertam o CO de forma controlada", explicou a investigadora a propósito do modo de funcionamento dos compostos. O próprio organismo produz monóxido de carbono, mas em quantidades insuficientes para destruir as células de bactérias.
Abre-se a porta a novos tratamentos
Na investigação agora apresentada foram utilizadas concentrações de CO superiores àquelas produzidas pelo organismo humano, mas não tóxicas. Com esta descoberta abre-se a porta a novos tratamentos e a novos antibióticos, capazes de contrariar a resistência que algumas bactérias desenvolveram em relação aos fármacos já existentes.
"A resistência de microrganismos patogénicos aos antibióticos clássicos é um dos maiores problemas que a medicina enfrenta, sendo responsável por um elevado número de mortes, especialmente em ambientes hospitalares" e "os compostos libertadores de CO poderão assim constituir uma nova geração de antibióticos", lê-se no comunicado.
Lígia Saraiva admitiu, no entanto, que "não há garantias que as bactérias não venham a desenvolver resistência a novos antibióticos que possam resultar desta descoberta". A investigadora do ITQB explicou que o trabalho que desenvolve no laboratório é "compreender o processo pelo qual a bactéria morre" e que o instituto não pode fazer testes relativos ao uso farmacológico dos compostos libertadores de CO em humanos.
No entanto, Lígia Saraiva adiantou que a empresa farmacêutica detentora da patente destes compostos pretende iniciar em breve os testes em animais. A descoberta da equipa de investigadores coordenada por Lígia Saraiva, e da qual também fazem parte Carlos Romão, João Seixas e Lígia Nobre, será publicada na edição de Dezembro da revista internacional "Antimicrobial Agents and Chemotherapy".
![]()
Portugueses descobrem que CO pode ser antibiótico
04.12.2007
Nova Geração de antibióticos? Uma equipa de investigadores do Instituto de Tecnologia Química e Biológica de Oeiras descobriu que o monóxido de carbono, pode conduzir a uma nova geração de antibióticos.
Os investigadores descobriram que o monóxido de carbono, apesar de ser um gás letal em doses elevadas, pode vir a dar origem a uma nova geração de antibióticos quando usado em doses baixas, controladas e toleradas pelo organismo.
A investigadora Lígia Saraiva conta que o grande interesse desses futuros antibióticos é que são uma novidade para as bactérias multiresistentes já habituadas aos antibióticos clássicos.
«É um modo de acção completamente distinto dos antibióticos normais, porque tem um composto de diferente base química. E isso é que é importante porque é uma grande batalha da comunidade cientifica», salienta a investigadora.
O estudos dos investigadores portugueses aponta para as seguintes conclusões: «Nós sujeitamos as bactéria a essa concentração de CO a ver se sobreviviam ou não. E verificamos que não só param o crescimento como morrem. Podiam ficar parada ou letárgicas, mas o CO tem mesmo capacidade para matar».
O estudo desta equipa de investigadores portugueses pode vir a resultar numa nova geração de antibióticos à base de monóxido de carbono. No entanto, Lígia Saraiva deixa um aviso:«Estamos a falar de quantidades baixas e controladas. O perigo do monóxido como gás tóxico mantém-se», salienta a cientista. É preciso, portanto, continuar a ter cuidado com lareiras, poluição e com o tabaco.
Awards
At international meeting on Helicobacter Infections : Best presentation prize
Oeiras, 24.07.12
After being selected for an oral presentation, the work “NorH, The First Nitric Oxide Reductase of Helicobacter pylori” by the Molecular Genetics of Microbial Resistance Lab was considered the Best Presentation Overall, among all poster and oral presentations presented at the 10th International Workshop on Pathogenesis and Host Response in Helicobacter Infections, held in Helsingor, Denmark, this July. The presentation by post-doc Marta C. Justino reported the discovery and biochemical and pathophysiological characterization of this novel enzyme.
At international meeting on Biometals: Best poster award
Oeiras, 24.07.12
The work by the Molecular Genetics of Microbial Resistance Lab, on the “Influence of stress conditions on iron homeostasis of Staphylococcus aureus”, by Lígia S. Nobre and co-authors, Catarina C. Godinho and Lígia M. Saraiva, received the best poster prize at the International Biometals 2012 meeting, which took place last week at Vrije Universiteit Brussel (VUB), Belgium.
Biosynthesis of bacterial tetrapyrroles on focus at Gordon Conference: Best poster award
Oeiras, 13.09.10
 |
The importance of NO: Young scientist awarded at “Congrès 2009 du Club NO”
Oeiras, 12.11.09
|
Marta C. Justino, post-doc at the Molecular Genetics of Microbial Resistance Lab, received the “Prix Jeunes Chercheurs” for her oral presentation “Ric proteins – A novel system for repair of nitrosatively and oxidatively damaged Fe-S clusters” during the “Congrès 2009 du Club NO” in Paris in October. |
 |
The Club NO France is an association which aims to further and promote all knowledge related to nitric oxide in both fundamental and applied sciences. It organises meetings and offers grants to exceptional students, post-docs and principal investigators. It is fast becoming the most useful resource for all nitric oxide researchers from all over the world. Among the 60 participants attending the “Congrès 2009 du Club NO", two “Prix Jeunes Chercheurs” were attributed by the Club NO Board Members.Ric proteins – A novel system for repair of nitrosatively and oxidatively damaged Fe-S clusters
Back to ITQB MGMR external page